Case study
Scaling AI-driven drug discovery with Entity Explorer
My role: Design Lead & Contributor
Project type: Enterprise tools, B2B
Company: BenevolentAI
My contributions: Design leadership, Data visualisation, Qualitative research, UX design, Visual design, Design system
Outcome: Accelerated access to AI models with reduced error and bias
The Challenge
BenevolentAI's mission to revolutionise drug development focused on early-stage discovery where costs of failure is relatively low but impact on clinical success is high.
The company built an extensive knowledge graph capturing current biological knowledge and deployed AI models to predict novel drug hypotheses.
The problem of evaluating AI outputs was already solved with the product called Target Triage.
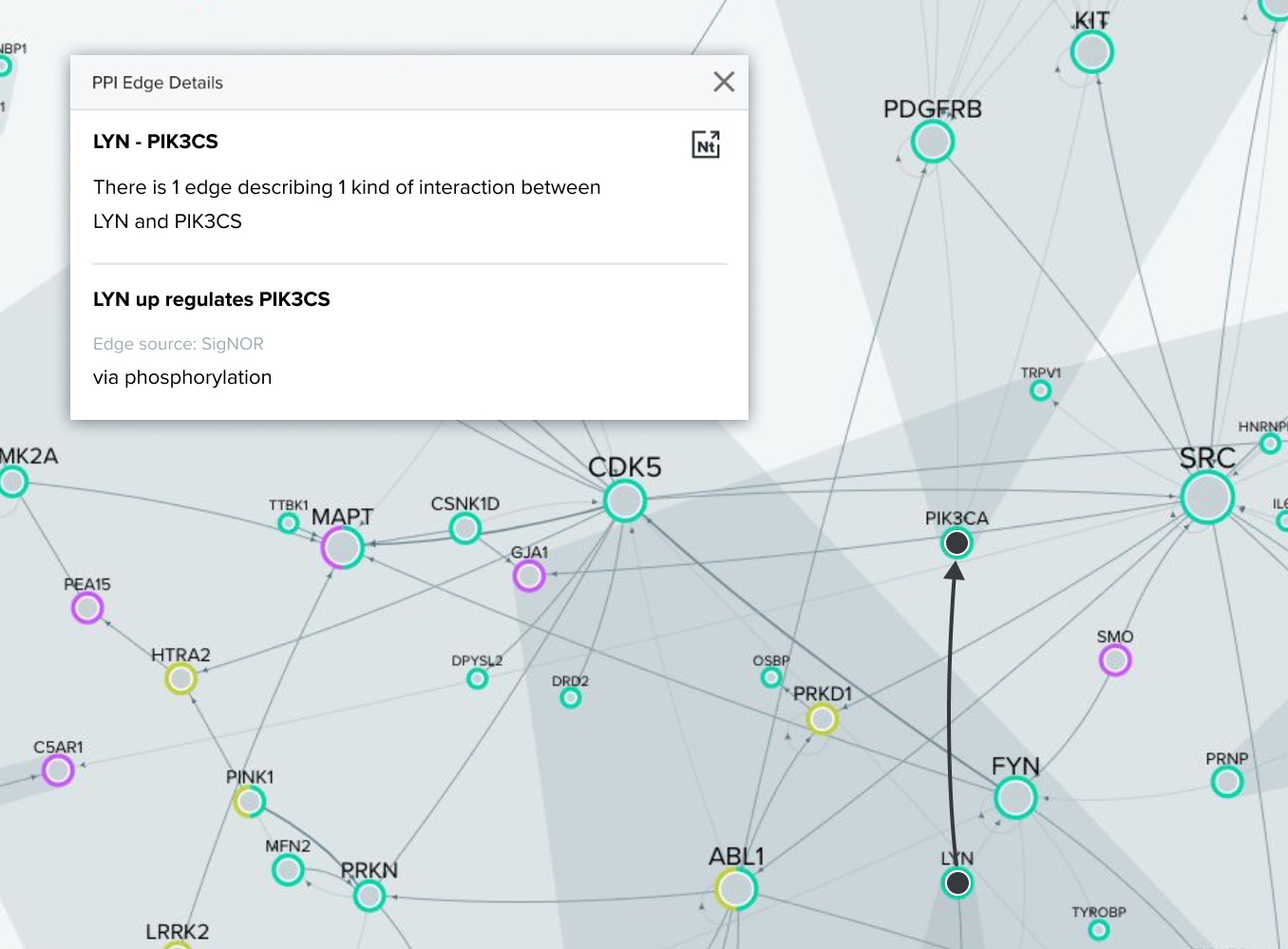
Entity selection is the process of identifying most relevant graph entities that will drive AI model predictions.
Best entities would be saved in the shortlist.
What remained was the challenge of selecting AI inputs — knowledge graph entities to drive AI models. Historically, entity selection was ran by bioinformaticians who provided multi-day data exploration services to drug discoverers and configured AI runs. This process was manual and slow, suffering from human bias and error, and often failing to capture the underlying molecular mechanism of disease.
To realise the benefits of AI-powered drug discovery, entity selection had to become faster, reliable, and precise.
My Approach
First, I led design-driven discovery to understand the problem and scope the solution in collaborate with product directors. Semi-structured interviews with drug discoverers and bioinformaticians, plus observing the legacy process, enabled first wireframes within days.
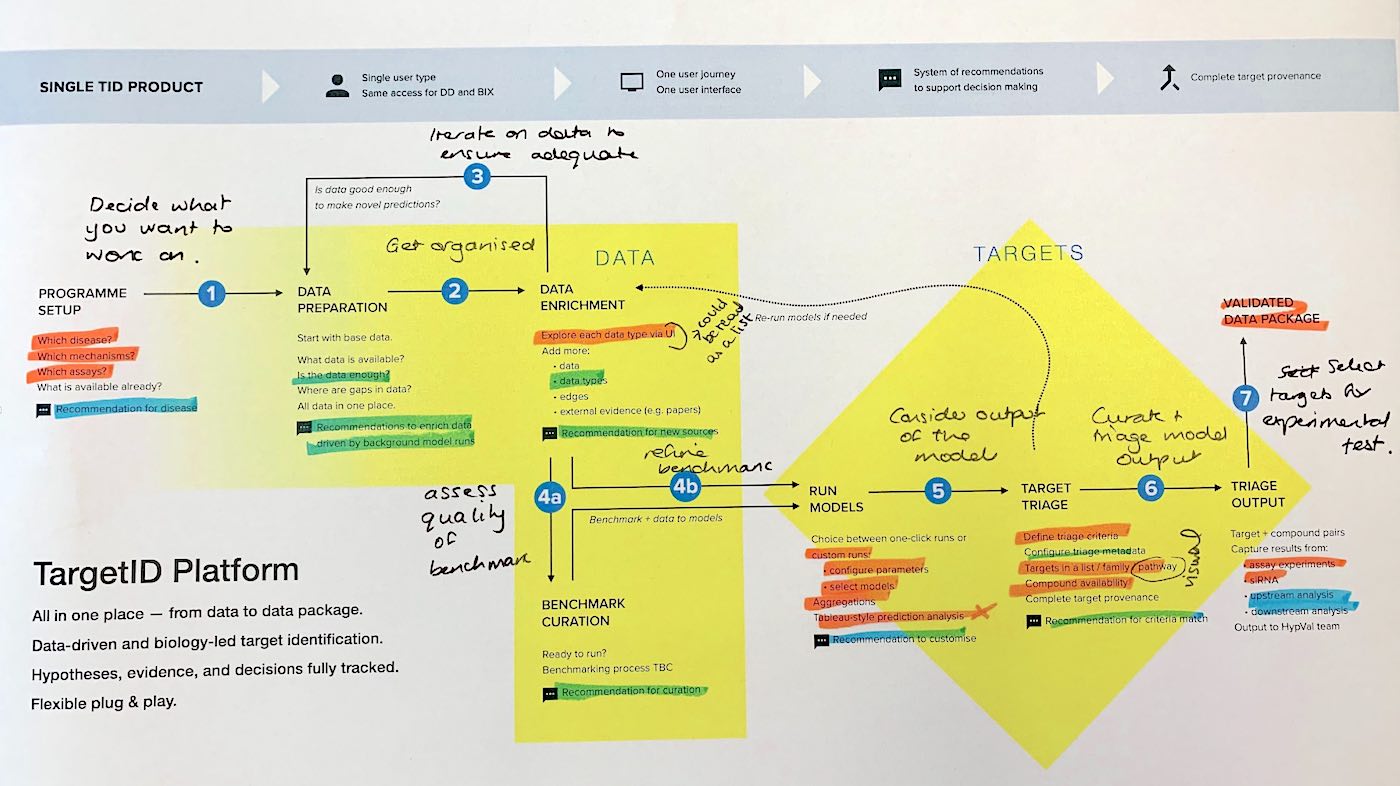
Stimulus-based interviews with internal users helped define Entity Explorer's vision within the end-to-end platform.
Wireframing and prototyping came next. I took the team through rapid iteration rounds, consulting with tech teams and validating with users.
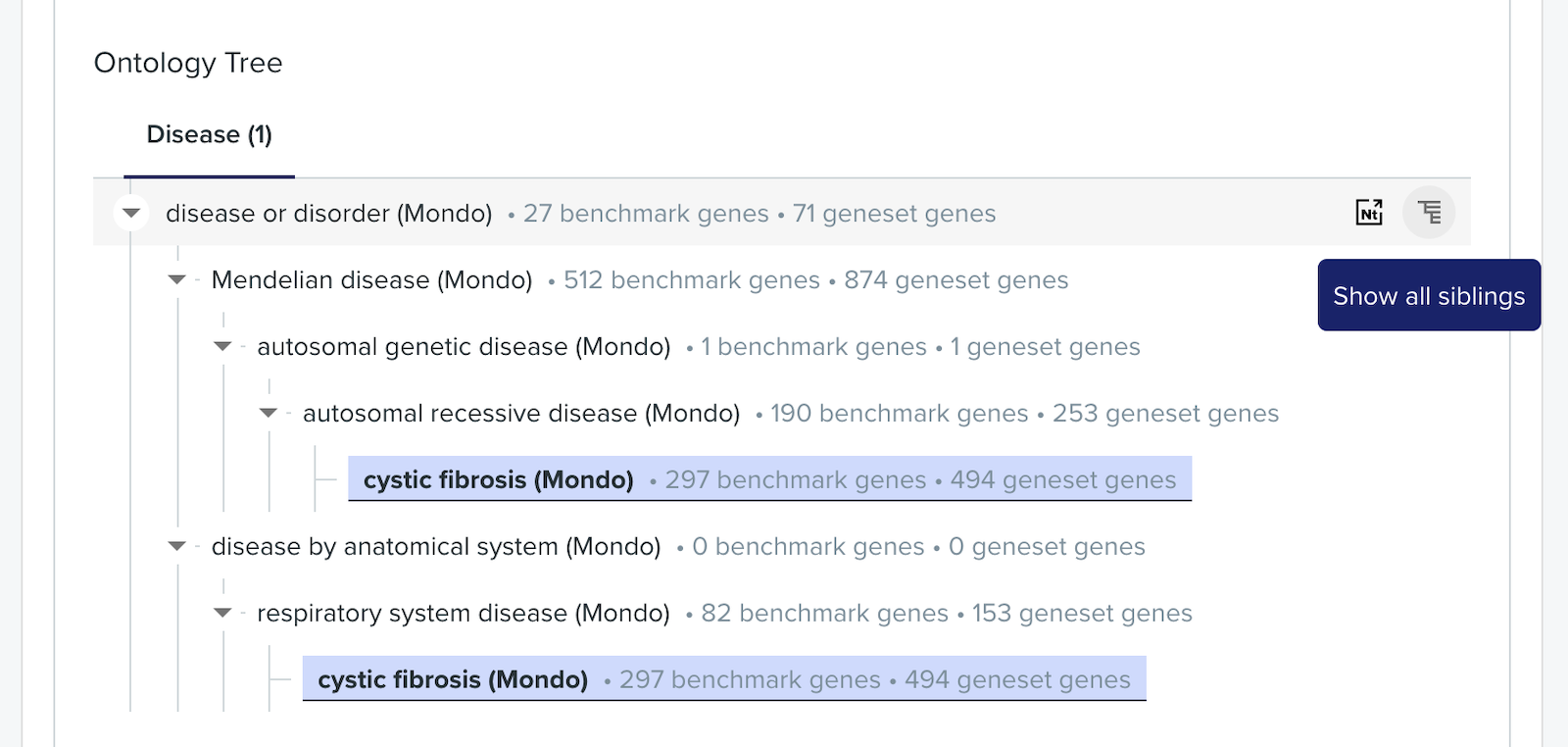
In parallel, I hired my design team. Once the product workflow was agreed, we split feature ownership between designers. I designed the Venn diagram, network visualisation, and the ontology tree.
In the background, we launched our Vue.js Design System.
It sped up development and enabled pair-working with engineers to take hand-drawn sketches straight into code (no hand-off).
The Solution
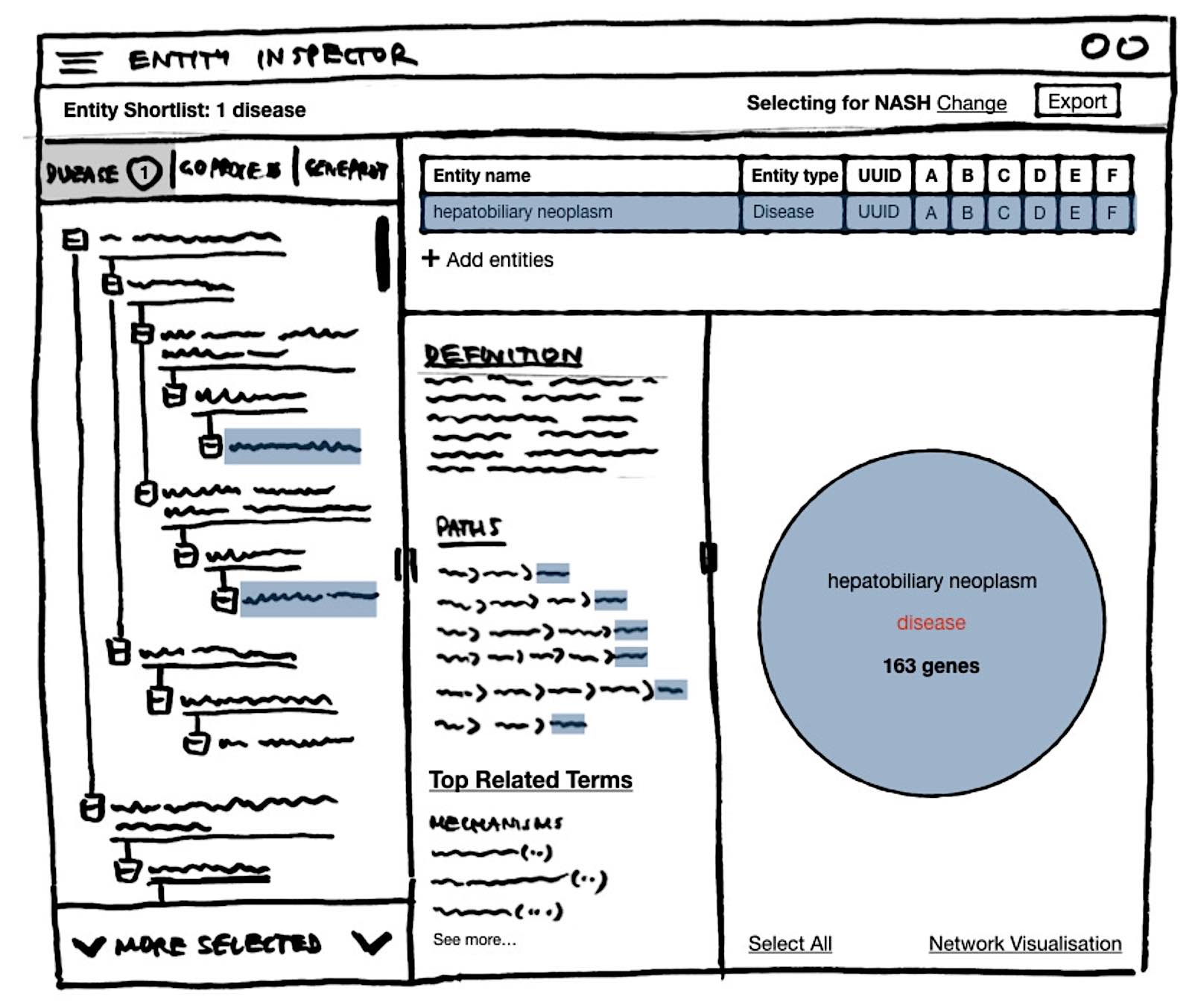
Entity Explorer is a web app that gives non-technical users ability to browse thousands of knowledge graph entities, compare entity benchmarks, explore ranked data-driven recommendations, visualise gene networks, and create entity shortlists for evaluation and AI model input.
The product forms the Benevolent Platform supporting end-to-end AI-powered early-stage drug discovery.
With other Benevolent Platform apps, Entity Explorer uses the Benevolent Design System, captures detailed user analytics (Mixpanel), and was deployed to BenevolentAI's commercial collaboration with AstraZeneca.
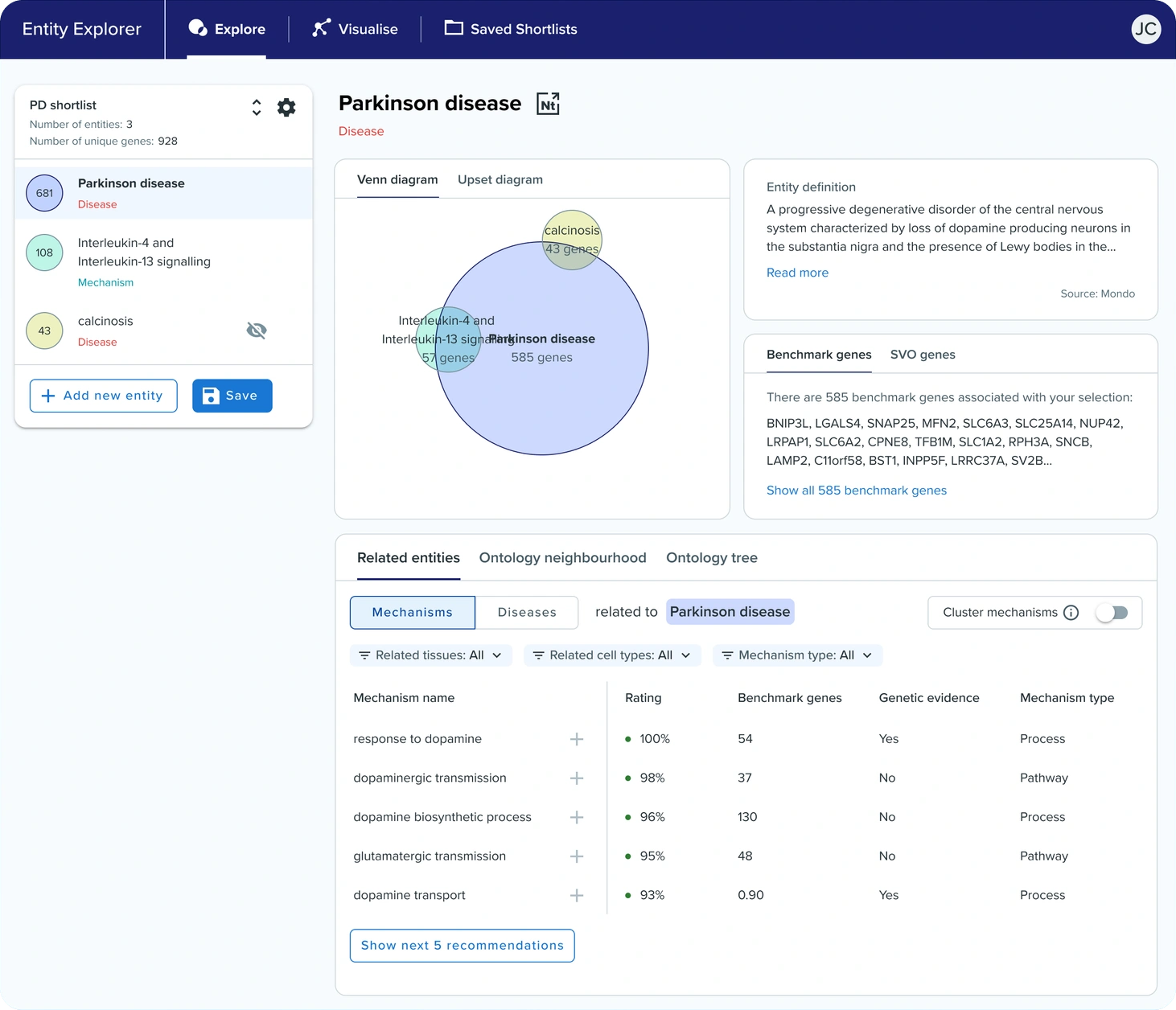
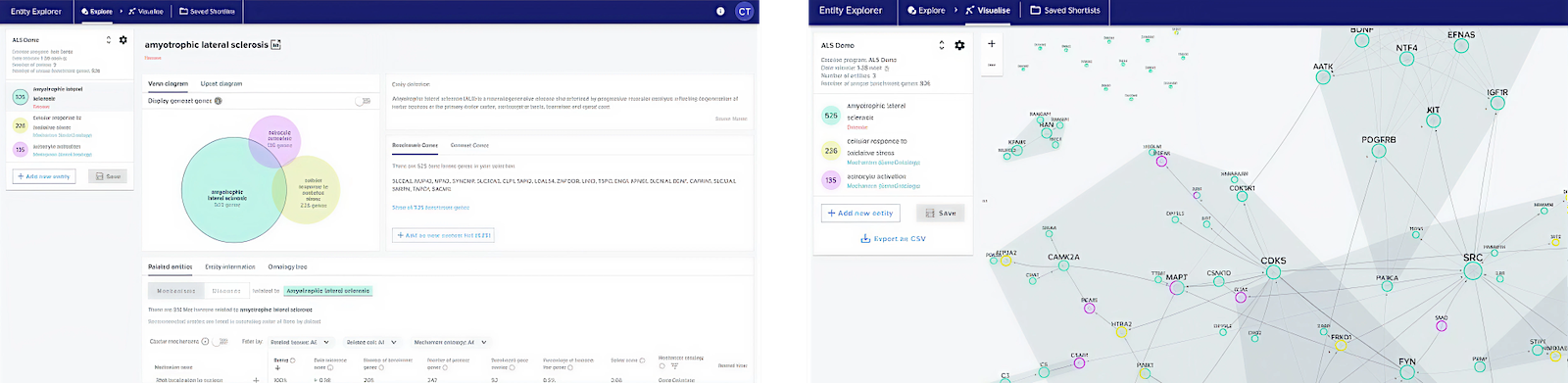

Entity Explorer gives access to entity exploration tools and a gene interaction network visualisation.
The Result
Entity Explorer gave drug discoverers self-service access to knowledge graph entities, letting them select AI inputs with no dependency on bioinformaticians (who could now focus on valuable R&D work).
This accelerated entity selection from days to hours and allowed BenevolentAI to build a pipeline of 26 programmes in 4 years (including 5 targets selected by AstraZeneca).
Entity Explorer made entity selection more reliable and robust. It eliminated the risk of human error, reduced bias, and helped teams understand disease biology to target underlying mechanisms with precision and confidence.
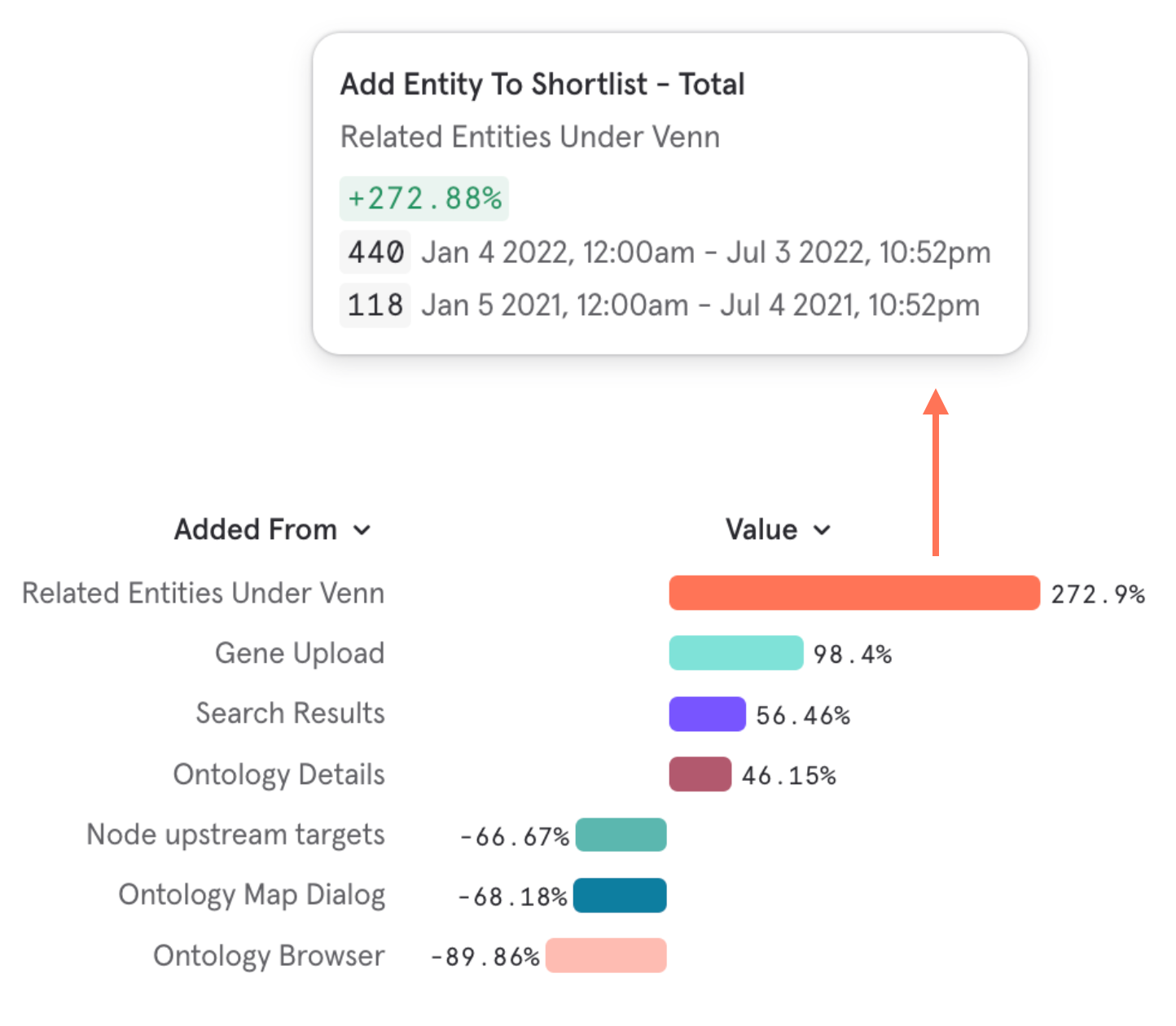
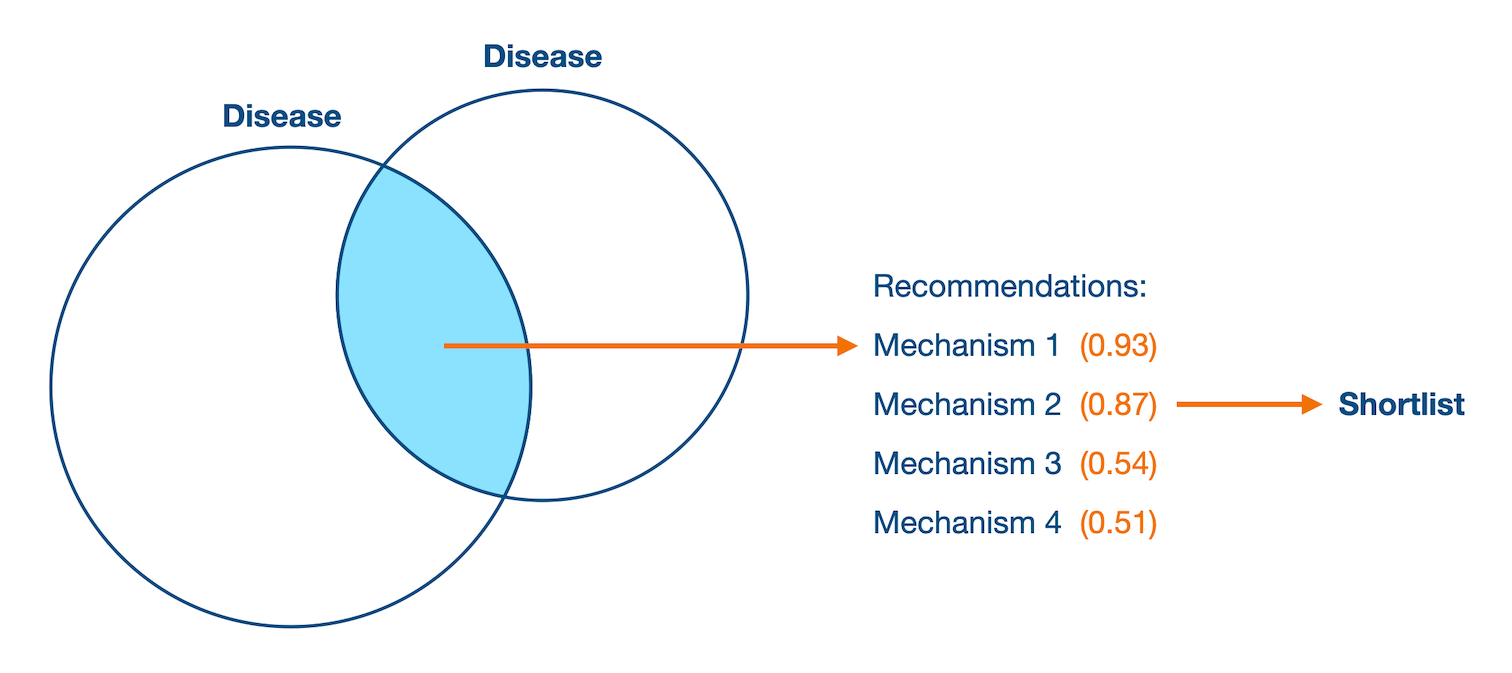
People bet on things they know well. This steered manual entity selection towards well-studied or trending terms.
To counter this bias, Entity Explorer provided entity recommendations matching user criteria. But they were underutilised.
I directed redesign work that grew the utilisation of data-driven recommendations by 270%.
The network visualisation was used in the BenevolentAI's drug discovery collaboration with AstraZeneca. AZ scientists praised its ease of use, interactive gene pathway exploration, and engaging interface.