Case study
Surfacing hidden patterns in data with Sequence Bundles
Video narrated by James Godwin
My role: Project & Design Lead
Project type: In-house R&D + External collaboration
Company: Science Practice
My contributions: Design leadership, Project management, Fundraising, Prototyping, Web development, Publishing
Outcome: New data patterns exposed with an award-winning visualisation tool
Website: Sequence Bundles
The Challenge
Bioinformatics is the cornerstone of modern life sciences. However, some of its early tools still struggle to provide answers to the problems investigated by modern researchers.
Popular tools for Multiple Sequence Alignment (MSAs) visualisation and analysis (such as Sequence Logo or pLogo) hide relational patterns in sequence data by aggregating information on a position-by-position basis. This means important dependencies and correlations between neighbouring positions or far-away locations are lost.
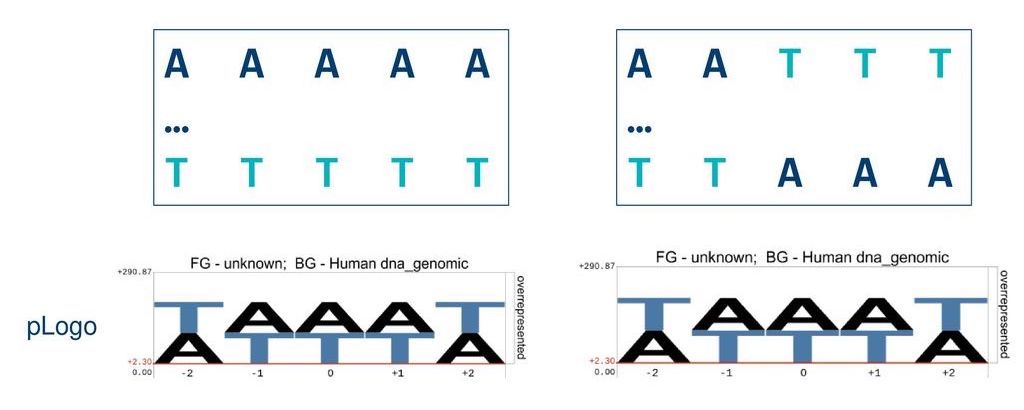
pLogo summarises data about each position in a sequence but hides important information about the sequence itself.
Here, two different sequences result in the same pLogo. We need new tools to expose sequential patterns in data.
How might we visualise sequence data differently to expose relational patterns in data and help scientists better understand biology, debug datasets, and drive scientific discovery further?
My Approach
My user-centred approach focused on target audiences in academia and industry. It helped secure interest from bioinformatics practitioners. I invited users to co-design, I prototyped, tested, and took the concept to conferences.
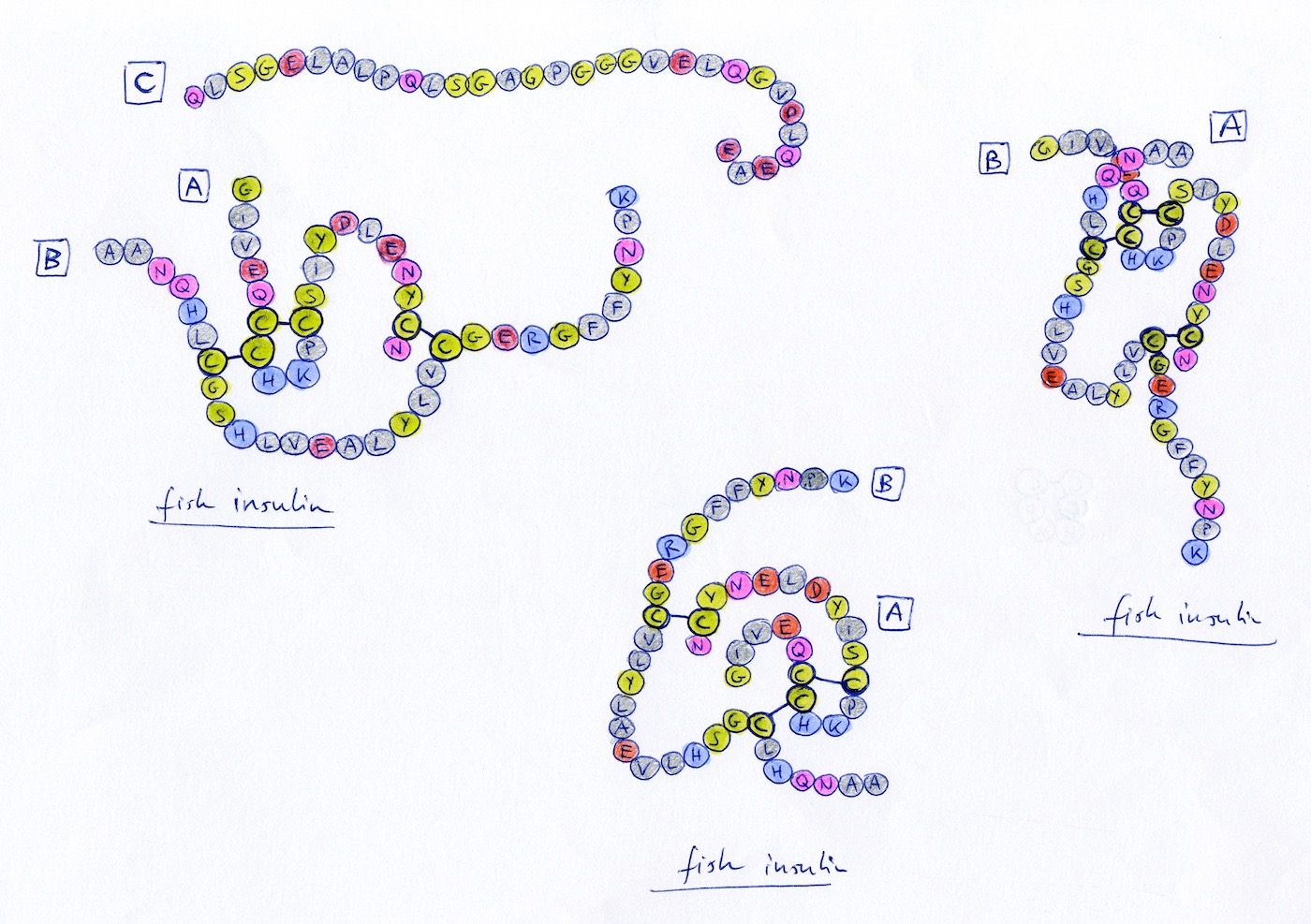
Early idea exploration and concept testing on paper. Focus is on leveraging sequence continuity to surface hidden patters.
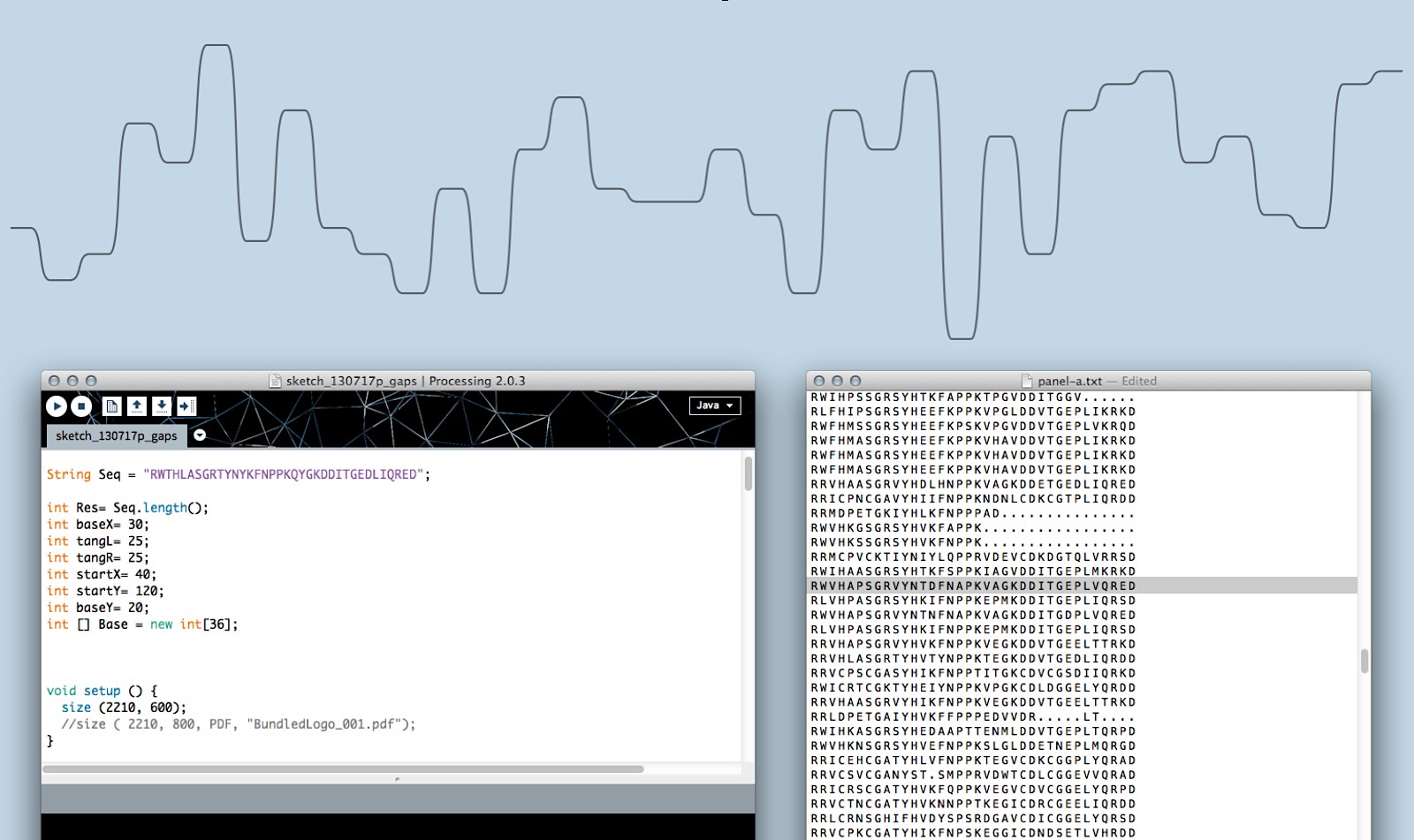
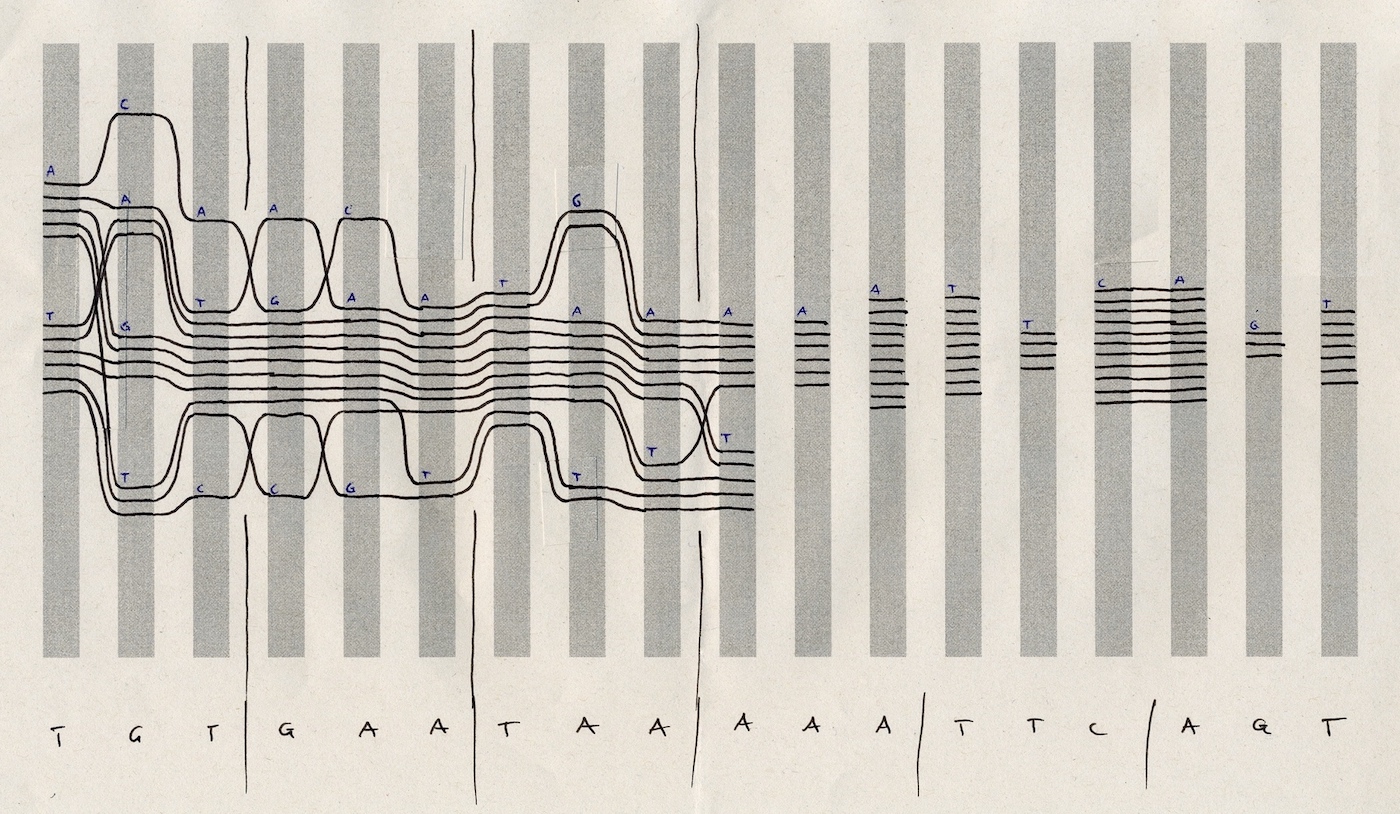
I prototyped in code using Processing.js to advance the concept and test at scale with real-life data.
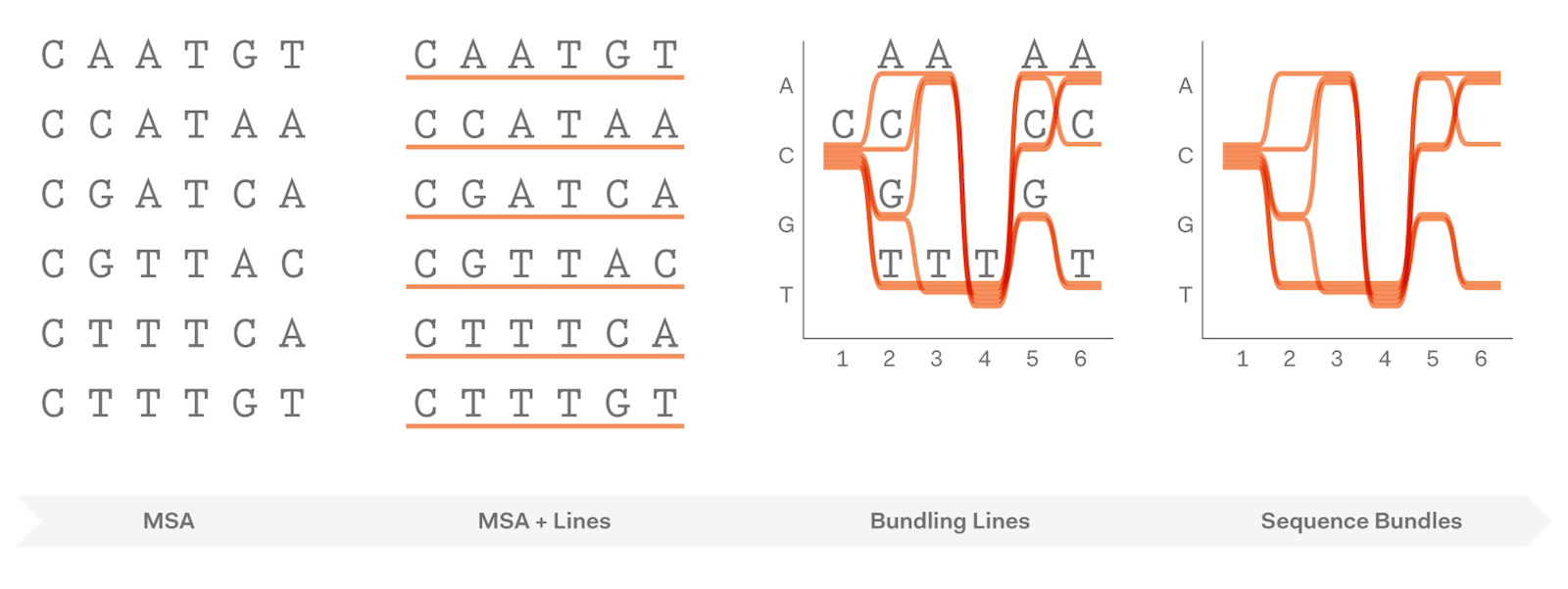
Each sequence is a line (Bezier curve). When letters in the sequence are the same (conserved), we bundle the lines together. Otherwise, we pull them apart.
This simple rule of visualisation exposes patterns in data that would otherwise remain hidden.
In 2014, I initiated a partnership between Science Practice and the Goldman Group at the EMBL-EBI to develop Sequence Bundles as a software tool. Our work was funded by a £33,000 InnovateUK grant that I secured.
I managed the project, led design work, supervised software development, contributed front-end code, and oversaw launch to the scientific community.
The Solution
Sequence Bundles is a novel MSA visualisation tool that exposes patterns in sequence data that would otherwise remain hidden. By preserving sequence continuity, the method provides meaningful visualisation that makes it possible to observe important residue correlations and uncover hidden sequence motifs.
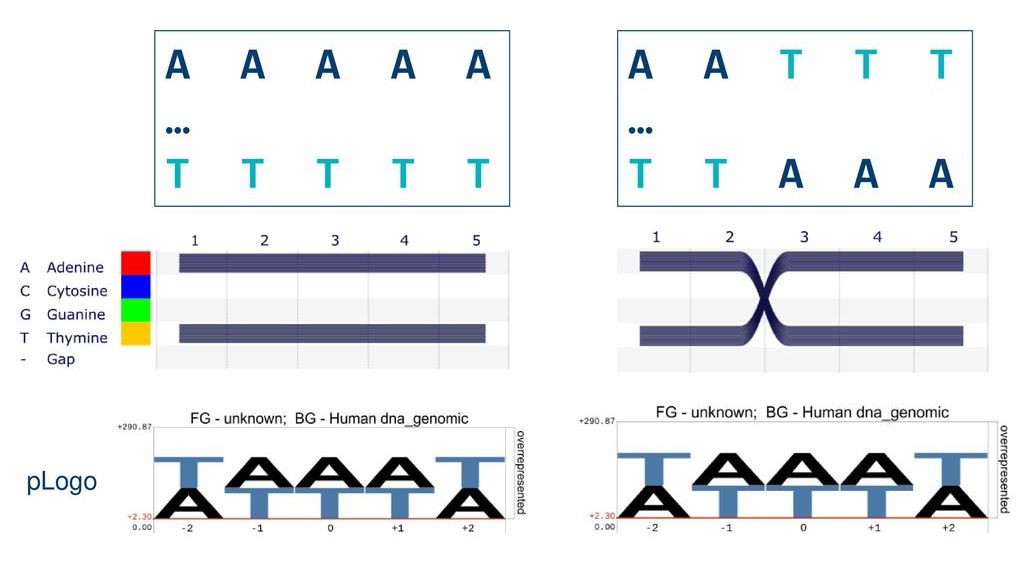
Sequence Bundles overcomes the limitations of a pLogo by visualising relational sequence motifs.
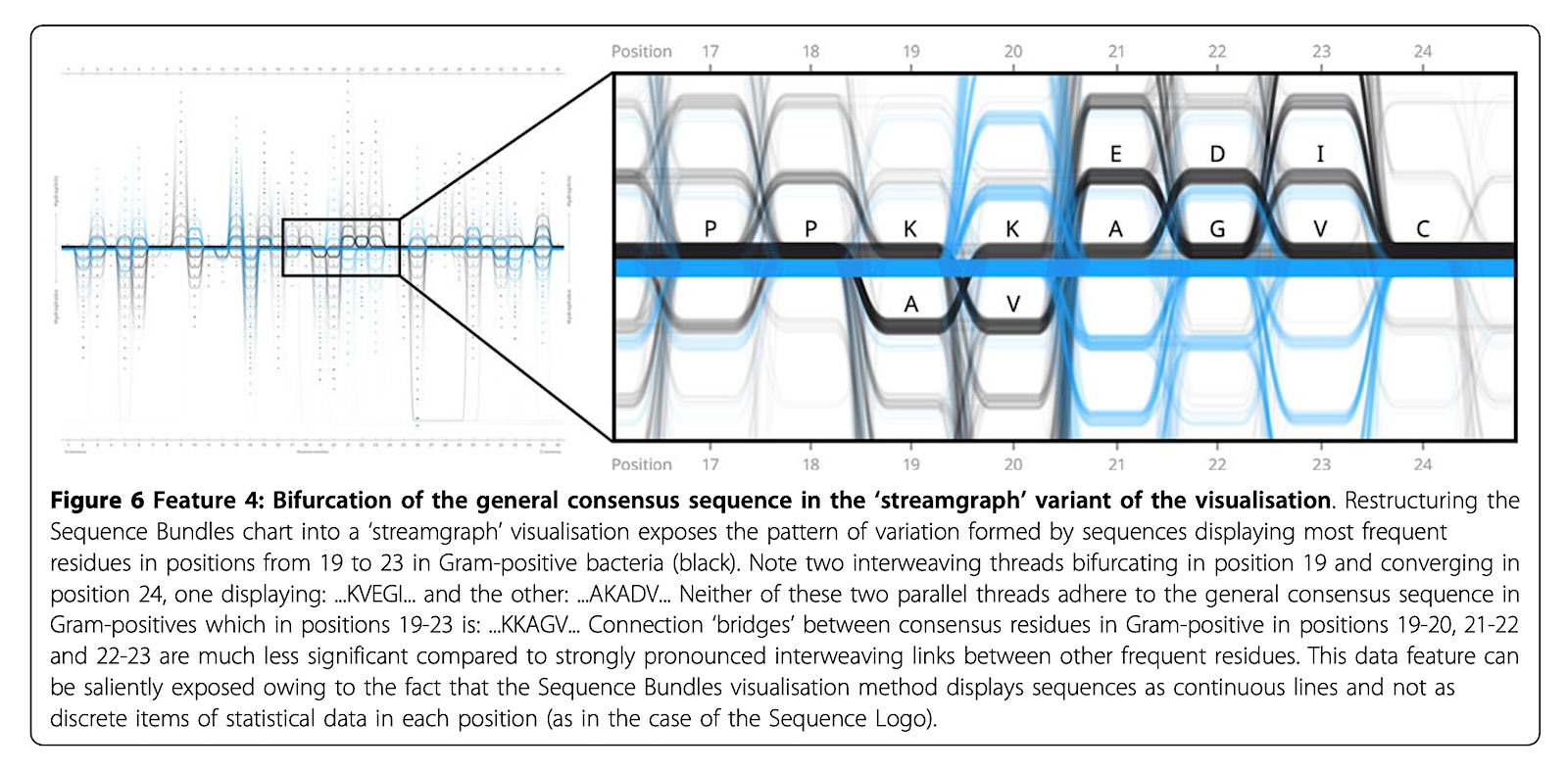
I found new patters in MSA data with Sequence Bundles. For example, a bifurcation in the general consensus sequence in positions 19–23 cannot be easily found using traditional methods (Figure 6).
Data source: BioVis 2013
Sequence Bundles was launched as a visualisation engine in combination with other bioinformatics analysis tools in a desktop app called Alvis, and as a quick-access web app online.
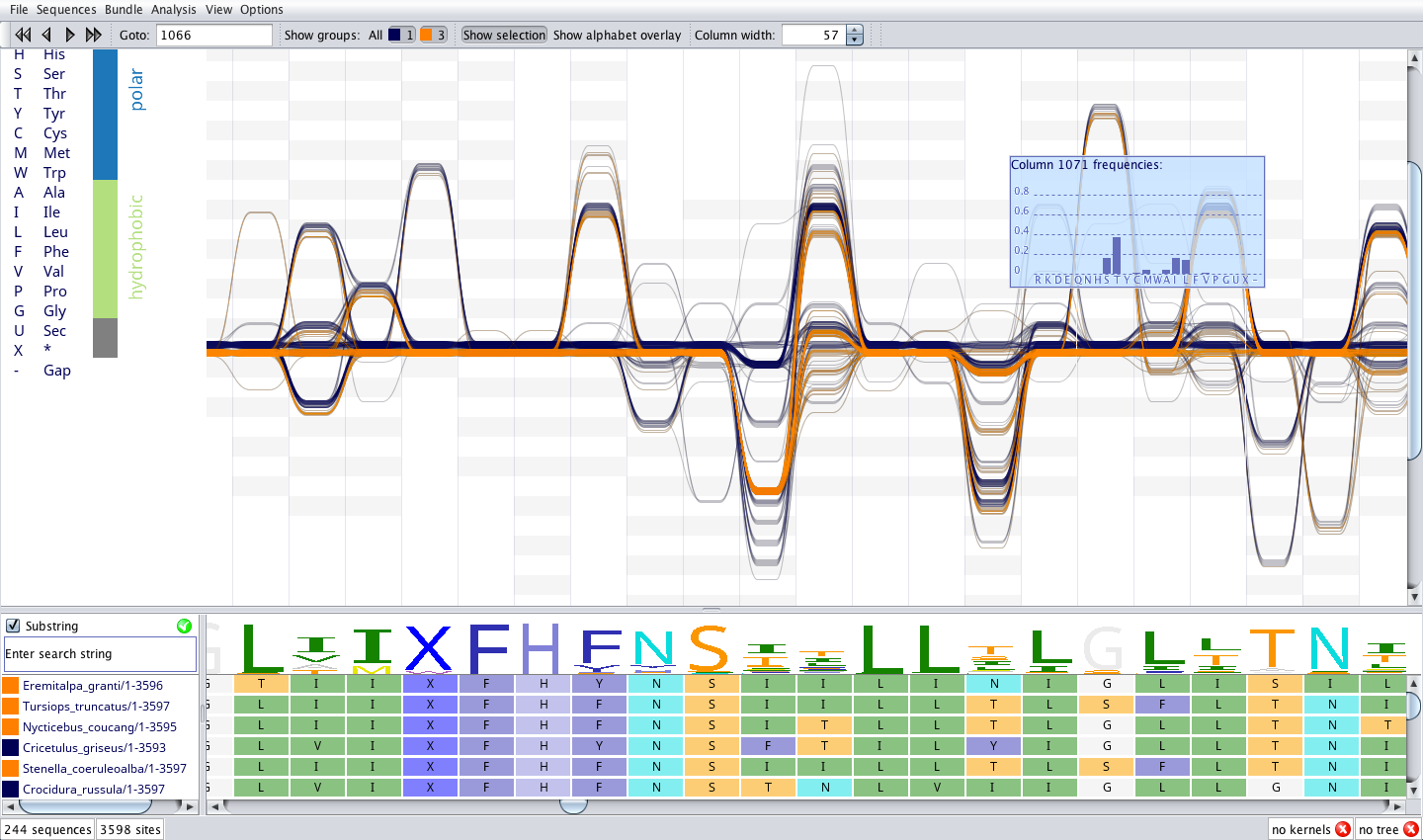
Alvis desktop app. It uses Sequence Bundles as its central visualisation tool. I led closed beta launch for Alvis. Later it was open-sourced by EMBL-EBI.
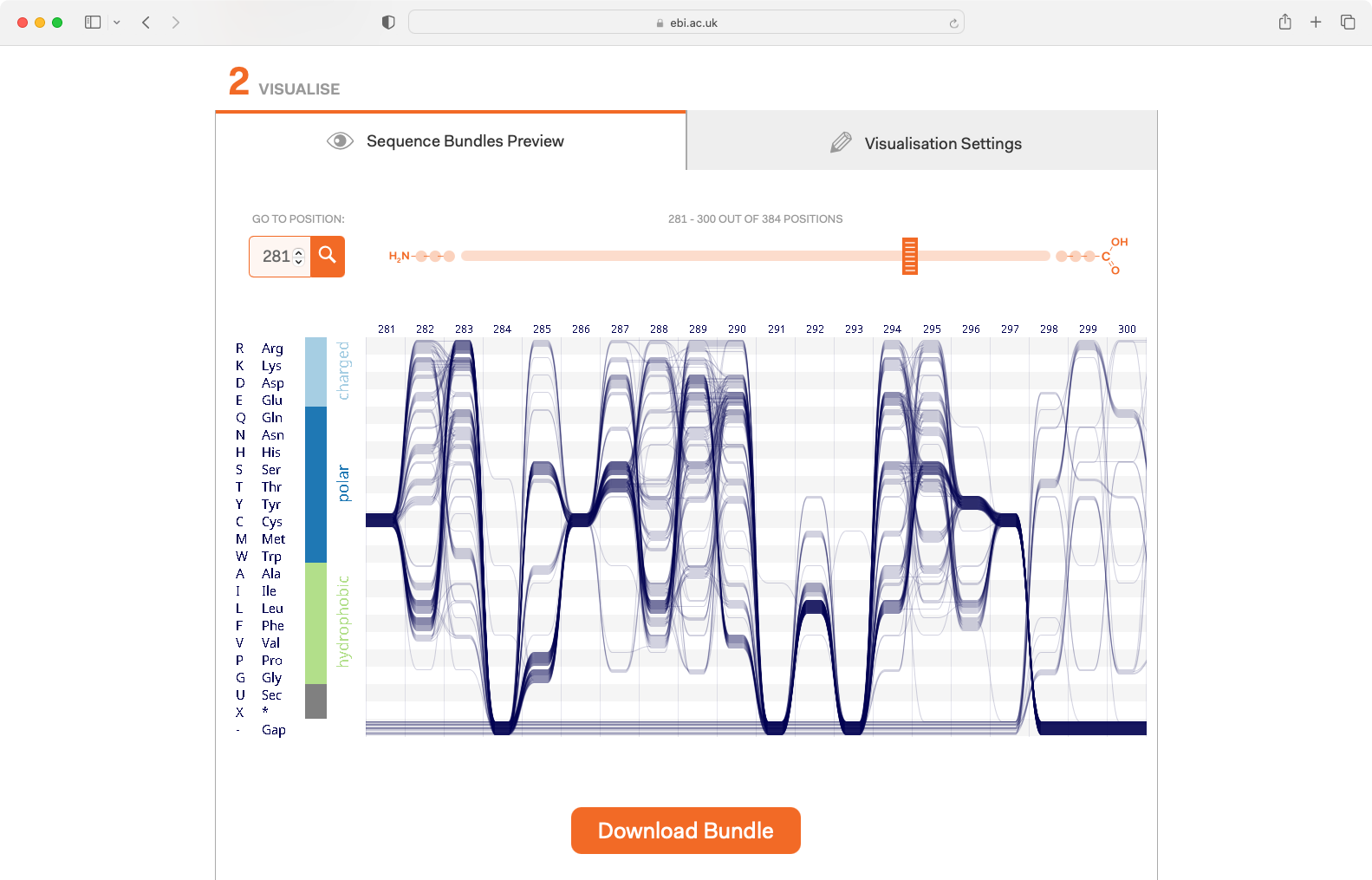
Sequence Bundles web app. I designed and co-developed the frontend. Backend service built in partnership with the Goldman Group at EMBL-EBI.
The Result
Sequence Bundles was published in peer-reviewed journals and won industry recognition.
To date, it received 27 academic citations, praise from bioinformatics practitioners, and industry awards including shortlist for the Kantar Information is Beautiful Awards 2016.
Sequence Bundles was a self-initiated science innovation project, funded by myself, Science Practice, and the InnovateUK grant.

User feedback shared in close beta by Meng Xu, Director of Bioinformatics at Hampton Creek.
I wrote the story of designing Sequence Bundles in more detail in this post on the Science Practice blog.